Team:Tokyo Tech/Experiment/C12
From 2012.igem.org
Contents |
Construction of the 3OC12HSL-dependent 3OC6HSL production module
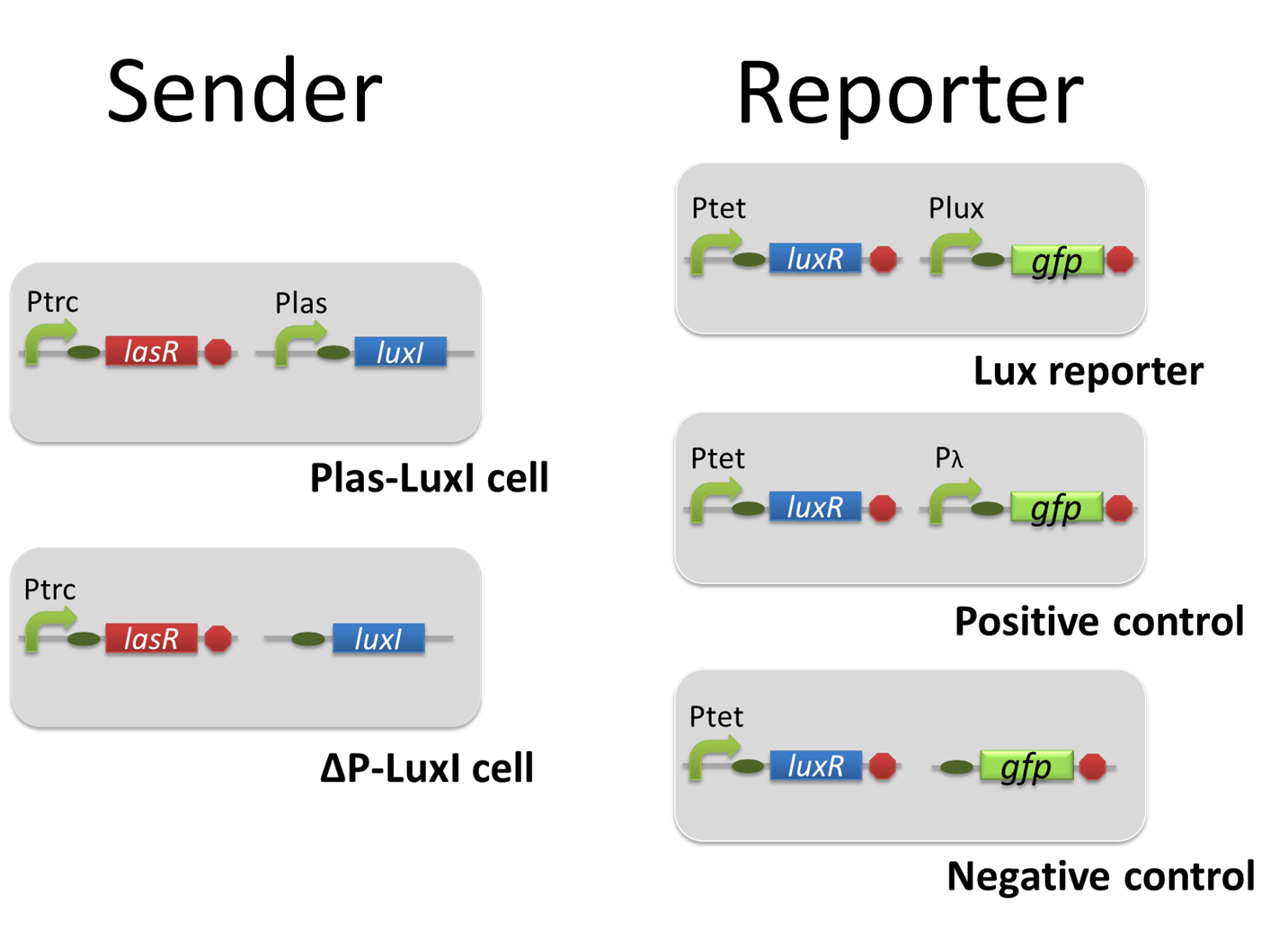
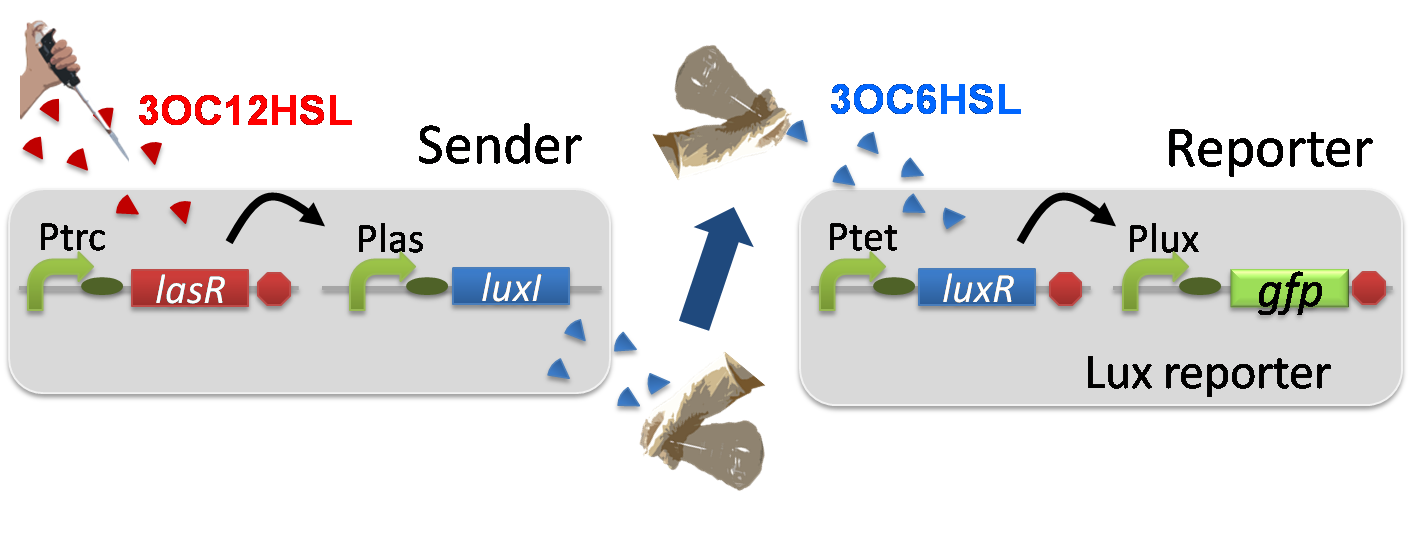
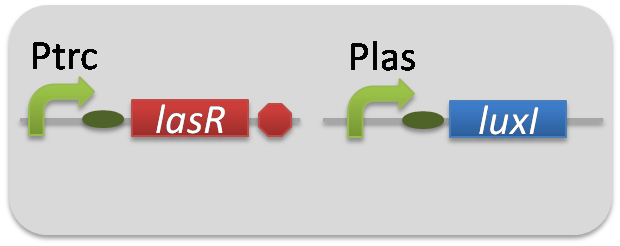
For construction of the 3OC12HSL-dependent 3OC6HSL production module, we firstly constructed a new part Plas-LuxI ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K934012 BBa_K934012]
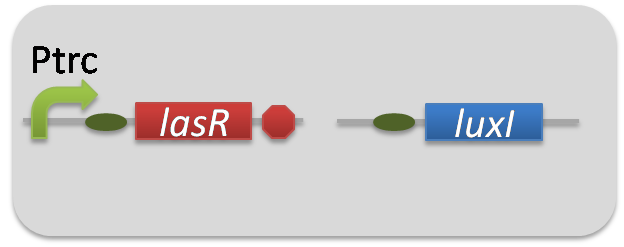
). Plas-LuxI cell is an engineered E.coli that contains a 3OC12HSL-dependent LuxI generator and a constitutive LasR generator. As the 3OC12HSL-dependent LuxI generator, we constructed a new Biobrick part Plas-LuxI ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K934012 BBa_K934012]
)by combining Plas promoter ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K649000 BBa_K649000]
). and LuxI ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K081008 BBa_K081008]
). As a constitutive LasR generator, we used Ptrc-LasR. By introducing Plas-LuxI and Ptrc-LasR into E.coli strain JM 2.300, we constructed Plas-LuxI cell.
Then we performed a reporter assay by using Lux reporter cell to characterize the function of Plas-LuxI. As the negative control of 3OC6HSL production, we prepared 3OC6HSL non-producer cell (ΔP-LuxI cell) that contains, in addition to Ptrc-LasR, promoterless-LuxI ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K081008 BBa_K081008]) instead of Plas-LuxI (Fig2-1-3-1-5).
The ΔP-LuxI cell does not produce 3OC6HSL even though 3OC12HSL exist. The supernatants of the cultures of these modules were used as the inducer in the reporter assay (Fig2-1-3-1-6).
We prepared four conditions as follow.
E) Culture containing Plas-LuxI cell without 3OC12HSL induction
F) Culture containing Plas-LuxI cell with 3OC12HSL induction
G) Culture containing ⊿P-LuxI cell without 3OC12HSL induction
H) Culture containing ⊿P-LuxI cell with 3OC12HSL induction
Using the supernatant of the four culture conditions, we performed the reporter assay.
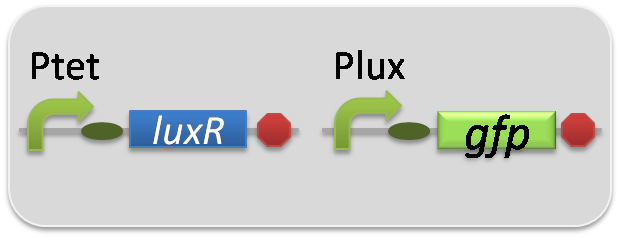
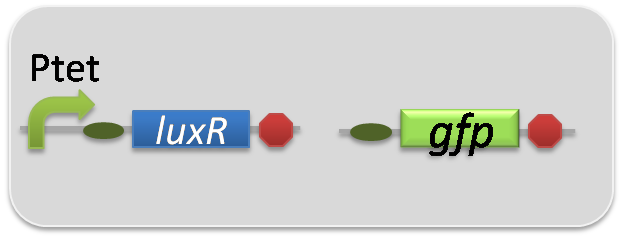
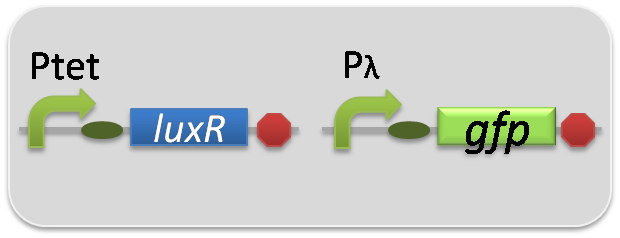
In the reporter assay, we used a Lux reporter strain that contains Ptet-LuxR and Plux-GFP ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K395100 BBa_K395100]). Also, a reporter cell that expresses GFP constitutively and a reporter cell that does not express GFP were used as the positive control and the negative control, respectively.
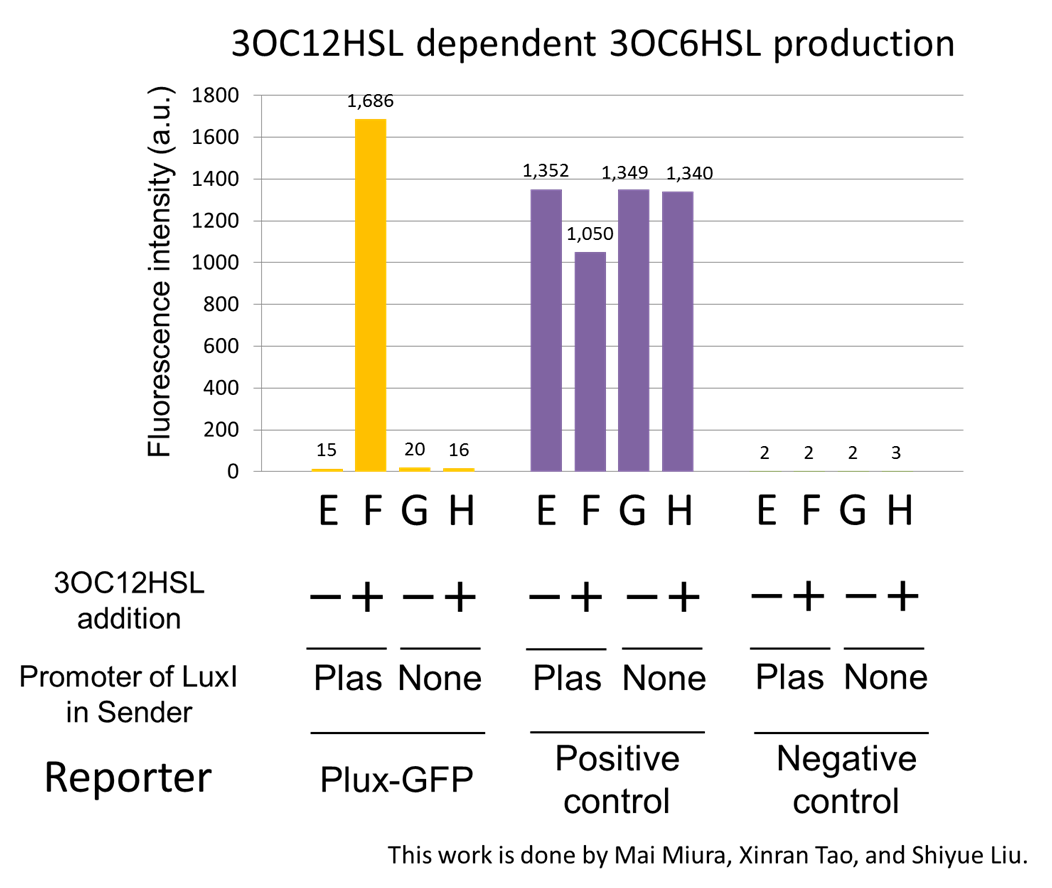
Fig2-1-3-1-7 shows fluorescence intensities by the reporter cells dependent on different conditions. Only when the supernatant of condition F was used, the fluorescence intensity of the Lux reporter cell increased while the supernatants of other three conditions did not affect. Comparing the results of the condition E and F, it can be said that with the induction of 3OC12HSL to Plas-LuxI, the fluorescence intensity of the Lux reporter cell increased by 112-folds. This result indicates that Plas-LuxI cell produced 3OC6HSL in response to 3OC12HSL induction by the function of Plas-LuxI ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K934012 BBa_K934012]) From this experiment, we confirmed that a new part Plas-LuxI ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K934012 BBa_K934012]) synthesized enough concentration of 3OC6HSL to induce the Lux reporter cell.
Materials & Methods
[Go to the project page "Construction of the 3OC12HSL-dependent 3OC6HSL production module"]
1.Construction
pSB6A1-Ptrc-LasR / pSB3K3-Plas-LuxI (JM2.300)…Plas-LuxI cell
pSB6A1-Ptrc-LasR / pSB3K3-ΔP-LuxI (JM2.300)…ΔP-LuxI cell
pSB6A1-Ptet-LuxR / pSB3K3-Plux-GFP (JM2.300)…Lux reporter cell
pSB6A1-Ptet-LuxR / pSB3K3-ΔP-GFP (JM2.300)…negative control
pSB6A1-Ptet-LuxR / pSB3K3-pλ-GFP (JM2.300)…positive control
2.Strain
JM2.300
Protocol
[Go to the project page "Construction of the 3OC12HSL-dependent 3OC6HSL production module"]
1. collect liquid culture
1.1 Prepare overnight culture of inducer cell at 37°C for 12hours.
1.2 Take 30μl of the overnight culture of inducer cell into LB(3ml) + antibiotics (Amp 50μg/ml + Kan 30μg/ml).(→fresh culture)
1.3 Incubate the flesh culture of inducer cell until the observed OD600 reaches around 0.50. Centrifuge the cell at 5000g, 25°C, 1 min, suspend it with 1ml LB + antibiotics (Amp 50μg/ml).
1.4 Take 30μl cell suspensions into LB(3ml) + antibiotics (Amp 50μg/ml + Kan 30μg/ml) + 5μM 3OC6HSL(3μl) and LB(3ml) + antibiotics (Amp 50μg/ml + Kan 30μg/ml) + DMSO(3μl).
1.5 Incubate the 3OC12HSL producer cells for another 4 hours at 37°C.
1.6 Centrifuge the 3OC12HSL producer cells at 9000g, 4°C, 1 min, and filter the cultured cells.
1.7 Dilute the filtrate by LB + antibiotics (Amp + Kan) in 1:30.
2 Reporter assay
2.1 Prepare overnight culture of reporter cell at 37°C for 12hours. 2.2 Take 30μl of the overnight culture of reporter cell into LB(3ml) + antibiotics (Amp 50μg/ml + Kan 30μg/ml). (→fresh culture)
2.3 Incubate the flesh culture of reporter cell until the observed OD600 reaches around 0.50, and gather the supernatant of culture of inducer cell.
2.4 Centrifuge the reporter cell at 5000g, 25°C, 1 min, and take it into LB + antibiotics (Amp 50μg/ml + Kan 30μg/ml).
2.5 Add 30μl samples of process 2.4 to filtrate + LB(3ml) + antibiotics (Amp 50μg/ml + Kan 30μg/ml) from process 1.7
| 3OC12HSL | Plas |
| + | + |
| - | + |
| + | - |
| - | - |
2.6 Induction of reporter cell for 4 hours at 37°C.
2.7 Flow cytometer measurements for GFP expression of reporter cell.
 "
"